Usage
Kanapy’s CLI
Kanapy provides some basic command line instructions to manage the installation. The
$ kanapy --help command
details this as shown:
Note
Make sure that you are within the virtual environment created during the kanapy installation, as this environment contains the kanapy instalation and its required dependencies.
$ conda activate knpy
(knpy) $ kanapy --help
Usage: kanapy [OPTIONS] COMMAND [ARGS]...
Options:
--help Show this message and exit.
Commands:
copyExamples Creates local copy of Kanapy's example directory.
genDocs Generates a HTML-based reference documentation.
gui Start Kanapy's GUI
runTests Runs unittests built-in to Kanapy.
setupTexture Stores the user provided MATLAB & MTEX paths for texture.
Kanapy’s API (incomplete)
Under construction …
The central element of Kanapy are statistical microstructure descriptors, which are used to generate the microstructure geometry and its texture in the different Kanapy modules. For multiphase microstructures, each phase has its own statistical descriptors and the respective dictionaries and combined to a list.
Microstructure descriptors
An exemplary structure of the descriptor dictionary is shown below:
{
"Grain type":
"Elongated",
"Equivalent diameter":
{
"sig": 0.39,
"scale": 2.418,
"loc": 0.0,
"cutoff_min": 8.0,
"cutoff_max": 25.0
},
"Aspect ratio":
{
"sig": 0.3,
"scale": 0.918,
"loc": 0.0
"cutoff_min": 1.0,
"cutoff_max": 4.0
},
"Tilt angle":
{
"kappa": 15.8774,
"loc": 87.4178,
"cutoff_min": 75.0,
"cutoff_max": 105.0
},
"RVE":
{
"sideX": 85.9,
"sideY": 112.33076,
"sideZ": 85.9,
"Nx": 65,
"Ny": 85,
"Nz": 65
},
"Simulation":
{
"periodicity": "True",
"output_units": "mm"
},
"Phase":
{
"Name": "Simulanium",
"Number": 1,
"Volume fraction": 1.0
}
}
Sample dictionaries are provided in the examples shown below. Note that descriptors can
be stored and read from JSON files. In the following the essentail keywords:
Grain type, Equivalent diameter, Aspect ratio, Tilt angle, RVE, Simulation, Phase are described
Grain type(mandatory): Either “Elongated” or “Equiaxed”
Equivalent diameter(mandatory): takes in four arguments to generate a log-normal distribution for the particle’s equivalent diameter; they are the Normal distribution’s standard deviation and mean, and the minimum and maximum cut-off values for the diameter. The values should correspond to \(\mu m\) scale.
Aspect ratiotakes the mean and the standard deviation value value as input. If the resultant microstructure contains equiaxed grains then this field is not necessary.
Tilt anglekeyword represents the tilt angle of particles with respect to the positive x-axis. Hence, to generate a distribution, it takes in two arguments: the normal distribution’s mean and the standard deviation. If the resultant microstructure contains equiaxed grains then this field is also not necessary.
RVEtakes two types of input: the side lengths of the final RVE required and the number of voxels per RVE side length.
Simulationtakes in two inputs: A boolean value for periodicity (True/False) and the required unit scale (‘mm’ or ‘um’ = \(\mu m\)) for the output ABAQUS .inp file.
Phase(optional) Information about phase name, number and volume fraction.
Note
The user may choose not to use the built-in voxelization (meshing) routine for meshing the final RVE. Nevertheless, a value for the number of voxels per side has to be provided.
A good estimation for Nx, Ny & Nz value can be made by keeping the following point in mind: The smallest dimension of the smallest ellipsoid/sphere should contain at least 3 voxels.
The size of voxels should be the same along X, Y & Z directions (voxel_sizeX = voxel_sizeY = voxel_sizeZ). It is determined using: voxel size = RVE side length/Voxel per side.
Particles grow during the simulation. At the start of the simulation, all particles are initialized with null volume. At each time step, they grow in size by the value: diameter/1000. Since the maximum packing density of ellipsoides is about 65%, the full space filling structure is achieved during voxelization.
The input unit scale should be in \(\mu m\) and the user can choose between ‘mm’ or ‘um’ (= \(\mu m\)) as the unit scale in which output to the ABAQUS .inp file will be written.
Tutorials
Several examples in form of Python scripts and Jupyter notebooks come bundled along with the kanapy package. Each example describes a particular workflow and demonstrates the use of the geometry module and the texture module. Note, that the latter requires a complete Kanapy installation including MTEX, that can be setup with the ‘kanapy setupTexture’ command.
Notebooks
- The basic steps of using Kanapy to generate synthetic representative volume elements (RVE) are demonstrated. The RVE can be exported in standard FEA formats.
- An EBSD map aof a austenitic steel produced by additive manufacturing is analyzed with respect to statistical microstructure descriptors. The descriptors are the basis to build an RVE.
- A polycrystal with a second phase dispersed along the grain boundaries is generated.
Outdated CLI contents
The following sections refer mainly to CLI commands from previous Kanapy versions. To be updated …
Hence the examples are sub-categorized here under
two sub-sections: Geometry examples and Texture examples. A detailed description of these examples is presented here.
The two examples sphere_packing and ellipsoid_packing depict the different workflows
that have to be setup for generating synthetic microstructures with equiaxed and elongated
grains, respectively. And the two examples ODF_reconstruction and ODF_reconstruction_with_orientation_assignment
depict the workflows for reconstructing ODF from EBSD data and assigning orientations to RVE grains, respectively.
For a detailed understanding of the general framework of the packing simulations or the ODF reconstruction, please
refer to: Modeling.
Note
New examples must be created in a separate directory. It allows the kanapy modules an easy access to the json, dump and other files created during the simulation.
Workflows for sphere packing
This example demonstrates the workflow for generating synthetic microstructures with equiaxed grains. The principle involved in generating such microstructures are described in the sub-section Microstructure with equiaxed grains. With respect to the final RVE mesh, the user has the flexibility to choose between the in-built voxelization routine and external meshing softwares.
If external meshing is required, the positions and weights of the particles (spheres) after packing can be written out to be post-processed. The positions and weights can be read by the Voronoi tessellation and meshing software Neper for generating tessellations and FEM mesh. For more details refer to Neper’s documentation.
If the in-built voxelization routine is prefered, then kanapy will generate hexahedral element (C3D8) mesh that can be read by the commercial FEM software Abaqus. The Abaqus .inp file will be written out in either \(mm\) or \(\mu m\) scale.
> ms = knpy.Microstructure(descriptor=ms_elong, name=matname + '_' + texture + '_texture')
> ms.init_RVE() # initialize RVE including particle distribution and structured mesh
> ms.plot_stats_init() # plot initial statistics of equivalent grain diameter and aspect ratio
> ms.pack() # perform particle simulation to distribute grain nuclei in RVE volume
> ms.plot_ellipsoids() # plot final configuration of particles
> ms.voxelize() # assign voxels to grains according to particle configuration
> ms.plot_voxels(sliced=True) # plot voxels colored according to grain number
> ms.generate_grains() # generate a polyhedral hull around each voxelized grain
> ms.plot_grains() # plot polyhedral grains
> ms.plot_stats() # compared final grain statistics with initial parameters
After navigating to the directory where the input file stat_input.json is located, kanapy’s CLI
command genStats is executed along with its argument (name of the input file). It creates an exemplary
‘Input_distribution.png’ file depicting the Log-normal distribution corresponding to the input statistics defined in
stat_input.json. Modifications can be made to the input statistics based on this plot. Next the genRVE
command is executed to generate the necessary particle, RVE, and the simulation attributes, and it writes it
to json files. Next the pack command is called to run the particle packing simulation. This command looks
for the json files generated by genRVE and reads the files for extracting the information required for the
packing simulation. At each time step of the packing simulation, kanapy will write out a dump file containing
information of particle positions and other attributes. Finally, the neperOutput command (Optional) can be
called to write out the position and weights files required for further post-processing. This function takes
the specified timestep value as an input parameter and reads the corresponding, previously generated dump file.
By extracting the particle’s position and dimensions, it creates the sphere_positions.txt & sphere_weights.txt files.
Note
The
neperOutputcommand requires the simulation timestep as input. The choice of the timestep is very important. It is suggested to choose the time step at which the spheres are tightly packed and at which there is the least amount of overlap. The remaining empty spaces will get assigned to the closest sphere when it is sent to the tessellation and meshing routine. Please refer to Microstructure with equiaxed grains for more details.The values of position and weights for Neper will be written in \(\mu m\) scale only.
Note
For comparing the input and output statistics:
The json file
particle_data.jsonin the directory../json_files/can be used to read the particle’s equivalent diameter as input statistics.After tessellation, Neper can be used to generate the equivalent diameter for output statistics.
If the built-in voxelization is prefered, then the voxelize command can be called to generate the hexahedral mesh.
It populates the simulation box with voxels and assigns the voxels to the respective particles (Spheres). The
abaqusOutput command can be called to write out the Abaqus (.inp) input file. The workflow for this looks like:
$ conda activate knpy
(knpy) $ cd kanapy-master/examples/sphere_packing/
(knpy) $ kanapy genStats -f stat_input.json
(knpy) $ kanapy genRVE -f stat_input.json
(knpy) $ kanapy pack
(knpy) $ kanapy voxelize
(knpy) $ kanapy abaqusOutput
(knpy) $ kanapy outputStats
(knpy) $ kanapy plotStats
Note
The Abaqus (.inp) file will be written out in either \(mm\) or \(\mu m\) scale, depending on the user requirement specified in the input file.
For comparing the input and the output equivalent diameter statistics the
outputStatscommand can be called. This command writes the diameter values in either \(mm\) or \(\mu m\) scale, depending on the user requirement specified in the input file.The
outputStatscommand also writes out the L1-error between the input and output diameter distributions.The
plotStatscommand outputs a figure comparing the input and output diameter distributions.
Storing information in json & dump files is effective in making the workflow stages independent of one another. But the sequence of the workflow is important, for example: Running the packing routine before the statistics generation is not advised as the packing routine would not have any input to work on. Both the json and the dump files are human readable, and hence they help the user debug the code in case of simulation problems. The dump files can be read by the visualization software OVITO; this provides the user a visual aid to understand the physics behind packing. For more information regarding visualization, refer to Visualize the packing simulation.
Workflows for ellipsoid packing
This example demonstrates the workflow for generating synthetic microstructures with elongated grains. The principle involved in generating such microstructures is described in the sub-section Microstructure with elongated grains. With respect to the final RVE mesh, the built-in voxelization routine has to be used due to the inavailability of anisotropic tessellation techniques. The Module voxelization will generate a hexahedral element (C3D8) mesh that can be read by the commercial FEM software Abaqus.
$ conda activate knpy
(knpy) $ cd kanapy-master/examples/ellipsoid_packing/
(knpy) $ kanapy genStats -f stat_input.json
(knpy) $ kanapy genRVE -f stat_input.json
(knpy) $ kanapy pack
(knpy) $ kanapy voxelize
(knpy) $ kanapy abaqusOutput
(knpy) $ kanapy outputStats
(knpy) $ kanapy plotStats
The workflow is similar to the one described earlier for sphere packing. The only difference being, that the neperOutput
command is not applicable here. The outputStats command not only writes out the equivalent diameters, but also the
major and minor diameters of the ellipsoidal particles and grains.
Note
A good estimation for the RVE side length values can be made by keeping the following point in mind: The biggest dimension of the biggest ellipsoid/sphere should fit within the corresponding RVE side length.
For comparing the input and output equivalent, major and minor diameter statistics, the command
outputStatscan be called. Kanapy writes the diameter values in either \(mm\) or \(\mu m\) scale, depending on the user requirement specified in the input file.The
outputStatscommand also writes out the L1-error between the input and output diameter distributionsThe
plotStatscommand outputs figures comparing the input and output diameter & aspect ratio distributions.
Texture examples
Both examples ODF_reconstruction and ODF_reconstruction_with_orientation_assignment require MATLAB & MTEX to be
installed in your system. If your kanapy is not configured for texture analysis, please run the following command:
$ conda activate knpy
(knpy) $ kanapy setupTexture
Note
Your MATLAB version must be 2015 and above.
The required input files must be placed in the working directory from where the kanapy commands are run.
Workflow for ODF reconstruction
This example demonstrates the workflow for reconstructing ODF from experimental EBSD data. The principle involved
in generating the reduced ODF is described in the sub-section ODF reconstruction. Kanapy requires the EBSD data
saved as (.mat) file format. In this regard, an exemplary EBSD file (ebsd_316L.mat) is provided in the ../kanapy-master/examples/ODF_reconstruction/ folder.
$ conda activate knpy
(knpy) $ cd kanapy-master/examples/ODF_reconstruction/
(knpy) $ kanapy reduceODF -ebsd ebsd_316L.mat
After navigating to the directory where ebsd_316L.mat is located, kanapy’s CLI
command reduceODF is executed along with its argument (name of the EBSD (.mat) file). If kanapy’s
geometry module is executed already, then the number of reduced orientations are read directly. Else kanapy requests
the user to provide the number of reduced orientations required before calling the MATLAB ODF reconstruction algorithm.
Note
The EBSD (.mat) file is a mandatory requirement for the ODF reconstruction algorithm.
Note here the value of the kernel shape parameter (\(\kappa\)) is set to a default value of 0.0873 rad.
Alternatly, an initial kernel shape parameter (\(\kappa\)) can be specified as an user input (OR) the grains
estimated using MTEX can be provided as an input in the (.mat) file format. The value of \(\kappa\) must be in radians,
if user specified. Else if the grains (.mat) file is provided, then the optimum \(\kappa\) is estimated by kanapy using
the mean orientation of the grains. In this regard, an exemplary grains file (grains_316L.mat) is
provided in the ../kanapy-master/examples/ODF_reconstruction/ folder. The workflow for this looks like:
$ conda activate knpy
(knpy) $ cd kanapy-master/examples/ODF_reconstruction/
(knpy) $ kanapy reduceODF -ebsd ebsd_316L.mat -kernel 0.096
(OR)
(knpy) $ kanapy reduceODF -ebsd ebsd_316L.mat -grains grains_316L.mat
Note
The output files are saved to the
/mat_filesfolder under the current working directory.The output (.txt) file contains the following information: \(L_1\) error of ODF reconstruction, the initial (\(\kappa\)) and the optimized (\(\kappa^\prime\)) values, and a list of discrete orientations.
Additionaly kanapy saves the reduced ODF and the reduced orientations (.mat) files in this folder.
Kanapy writes a log file (
kanapyTexture.log) in the current working directory for possible errors and warnings debugging.
Workflow for ODF reconstruction with orientation assignment
This example demonstrates the workflow for reconstructing ODF from experimental EBSD data and then determining the optimal
assignment of orientations to RVE grains. The principle involved in optimal orientation assignment is described in the
sub-section ODF reconstruction with orientation assignment. In addition to the EBSD data, Kanapy requires
grain (.mat) file, and the grain boundary shared surface area information as input. In this regard, an exemplary
EBSD file (ebsd_316L.mat), and a grains file (grains_316L.mat) are provided in the
../kanapy-master/examples/ODF_reconstruction_with_orientation_assignment/ folder. It is important to note that
the grain boundary shared surface area file is created whilst generating an RVE by kanapy’s geometry module.
$ conda activate knpy
(knpy) $ cd kanapy-master/examples/ODF_reconstruction_orientation_assignment/
(knpy) $ kanapy genStats -f stat_input.json
(knpy) $ kanapy genRVE -f stat_input.json
(knpy) $ kanapy pack
(knpy) $ kanapy voxelize
(knpy) $ kanapy outputStats
(knpy) $ kanapy reduceODF -ebsd ebsd_316L.mat -grains grains_316L.mat -fit_mad yes
After navigating to the directory where the input file ebsd_316L.mat is located, generate an RVE by calling kanapy’s
geometry CLI commands: genStats, genRVE, pack & voxelize. To generate the shared surface area file,
run outputStats command. Kanapy will write a shared_surfaceArea.csv
file to the /json_files/ folder. This file contains the grain boundary shared surface area
information between neighbouring grains. Now, kanapy’s texture CLI command reduceODF can be called along with
its arguments (name of the EBSD, grains (.mat) files). The key -fit_mad must be used with this command to tell
kanapy that orientation assignment to grains is required. Since kanapy’s geometry module is executed already, kanapy recognizes
the number of reduced orientations required (=number of grains in the RVE). Else kanapy requests the user to provide
the number of reduced orientations required before calling the MATLAB functions.
Note
The EBSD, grains (.mat) files and the grain boundary shared surface file are mandatory requirements for the orientation assignment algorithm.
The
shared_surfaceArea.csvfile is generated by runningkanapy outputStats.
Additionally an optional input that can be provided is the grain volume information, which is used for weighting the
orientations after assignment and for estimating the ODF represented by the RVE. Kanapy also writes the grains volume file
(grainsVolumes.csv) to the /json_files/ folder, when the outputStats command is executed after RVE generation.
Note
The
grainsVolumes.csvfile lists the volume of each grain in the ascending order of the grain ID.Kanapy automatically detects the presence of the
shared_surfaceArea.csv&grainsVolumes.csvfiles, if they are present in the/json_files/folder.The output files are saved to the
/mat_filesfolder under the current working directory.The output (.txt) file contains the following information: \(L_1\) error of ODF reconstruction, \(L_1\) error between disorientation angle distributions from the EBSD data and the RVE, the initial (\(\kappa\)) and the optimized (\(\kappa^\prime\)) values, and a list of discrete orientations each with a specific grain number that it should be assigned to.
Additionaly kanapy saves the reduced ODF and the reduced orientations (.mat) files in this folder.
Kanapy writes a log file (
kanapyTexture.log) in the current working directory for possible errors and warnings debugging.
Visualize the packing simulation
You can view the data generated by the simulation (after the simulation
is complete or during the simulation) by launching OVITO and reading in
the dump files generated by kanapy from the ../sphere_packing/dump_files/ directory.
The dump file is generated at each timestep of the particle packing simulation. It contains
the timestep, the number of particles, the simulation box dimensions and the particle’s attributes
such as its ID, position (x, y, z), axes lengths (a, b, c) and tilt angle (Quaternion format - X, Y, Z, W).
The OVITO user interface when loaded, should look similar to this:
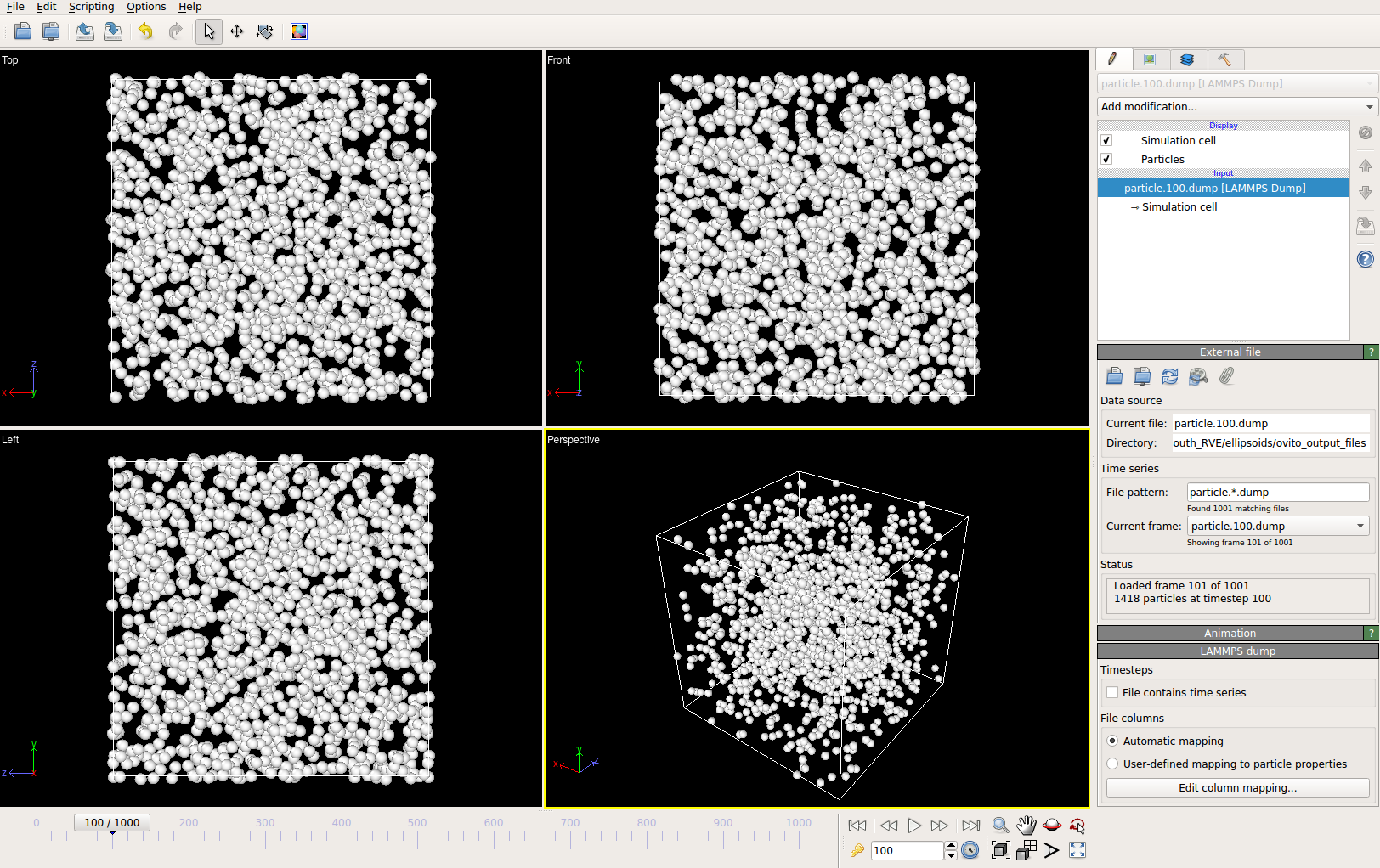
By default, OVITO loads the particles as spheres, this option can be changed to visualize ellipsoids.
The asphericalshapex, asphericalshapey, and asphericalshapez columns need to be mapped to
Aspherical Shape.X, Aspherical Shape.Y, and Aspherical Shape.Z properties of OVITO when
importing the dump file. Similarily, the orientationx, orientationy, orientationz, and
orientationw particle properties need to be mapped to the Orientation.X, Orientation.Y,
Orientation.Z, and Orientation.W. OVITO cannot set up this mapping automatically, you have
to do it manually by using the Edit column mapping button (at the bottom-right corner
of the GUI) in the file import panel after loading the dump files. The required assignment
and components are shown here:
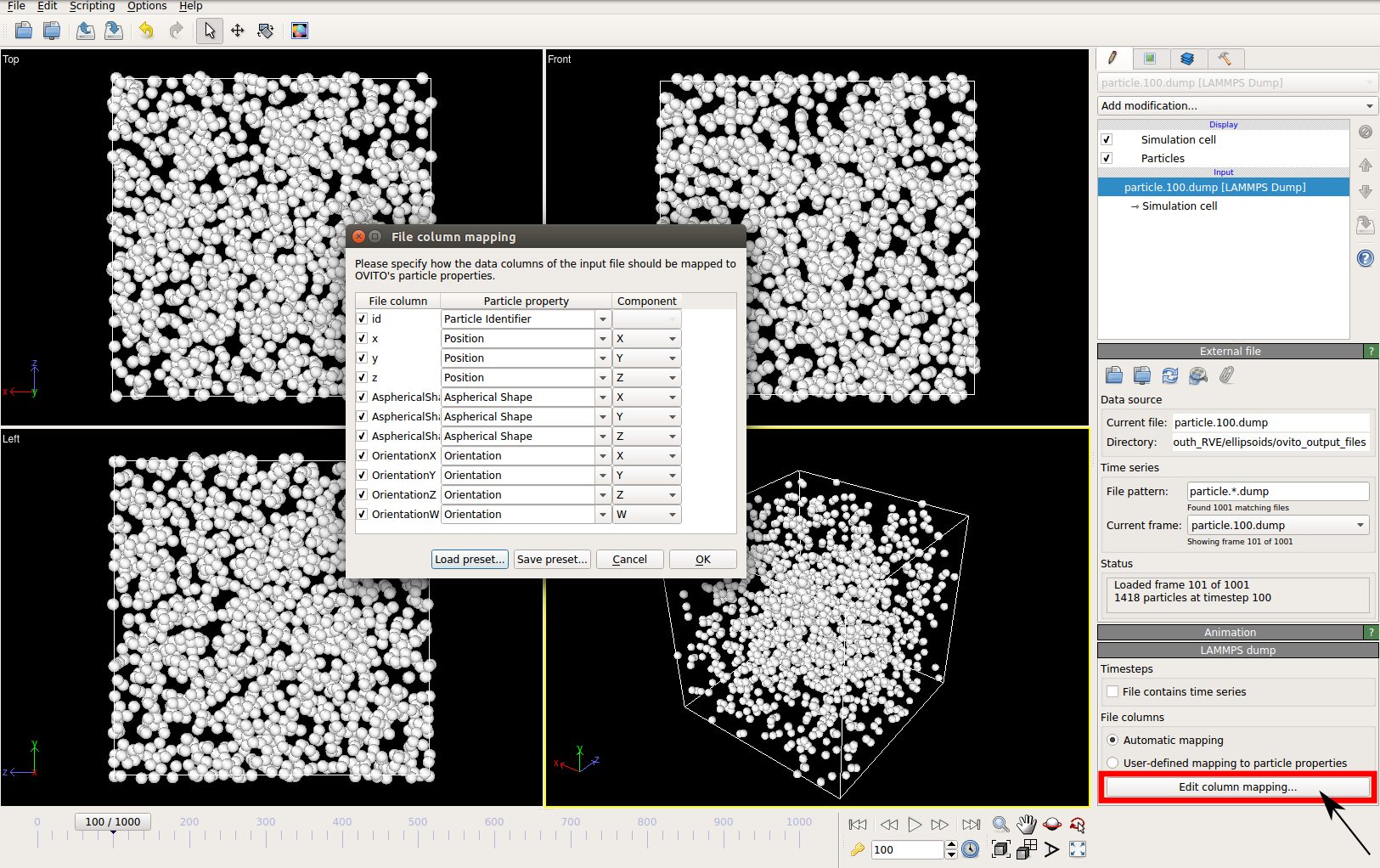
For further viewing customizations refer to OVITO’s documentation.